Molecular Docking Viewer
AI-Driven Design and In Silico Evaluation of Peptide Inhibitors For Dengue Virus Envelope Protein
This web application focuses on visualizing molecular docking results for protein–peptide interactions in a clear, interactive interface to support antiviral drug research.
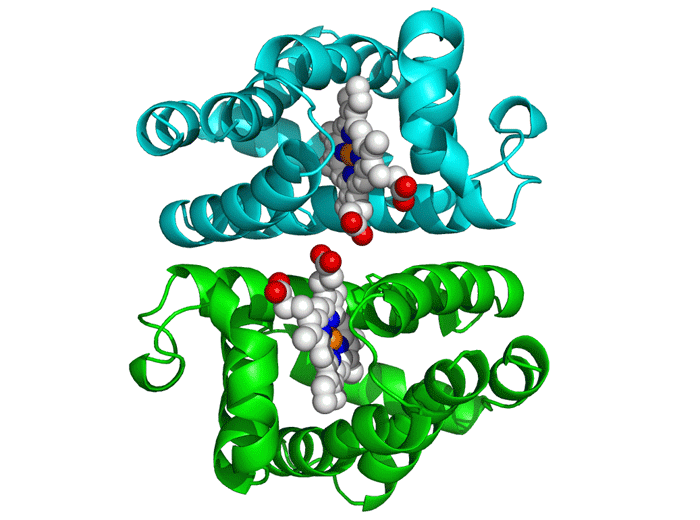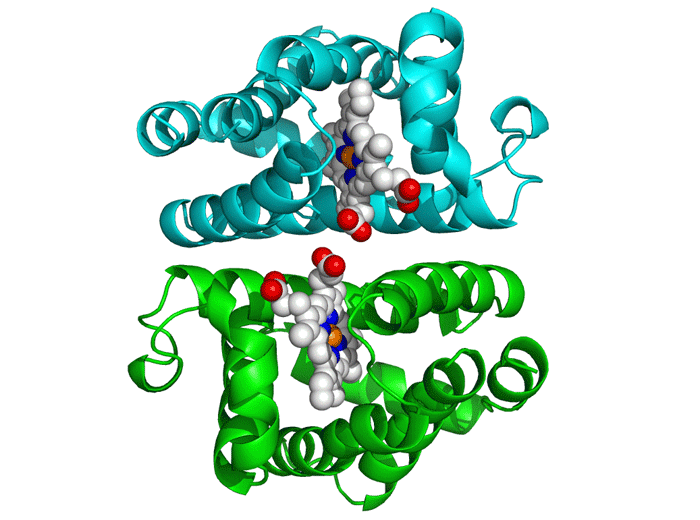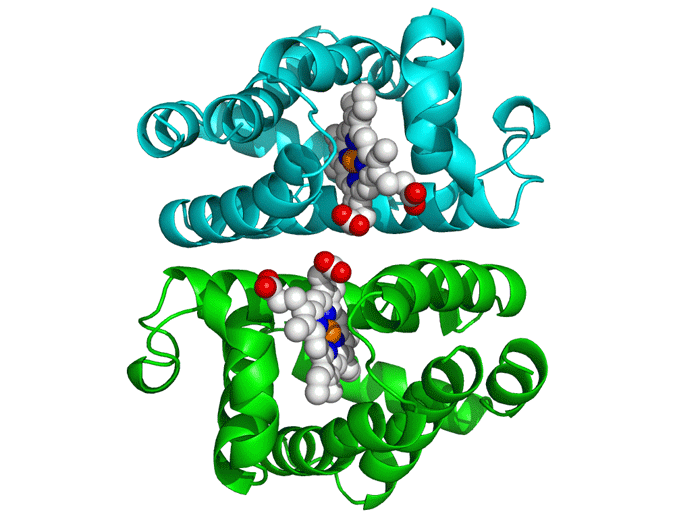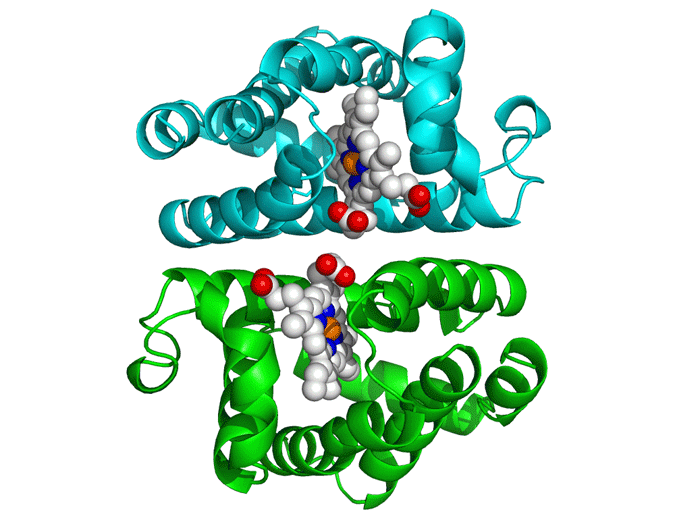
Project Objectives
Constructing antiviral peptide sequences that effectively inhibit and bind with the DENV E protein, utilizing deep learning models
Validating the predicted peptides using molecular docking to confirm their binding efficiency and stability within the target environment
Providing insights for further research development in the field of antiviral peptide discovery
Research Methodology
Peptide Generation
The initial phase involved generating novel peptide sequences using advanced machine learning techniques. We primarily utilized ProtT5, a Transformer-based protein language model, specifically its 8-bit quantized model to manage computational resource demands effectively.
Peptide sequences were carefully curated from established antiviral peptide databases including AVPDB and DRAVP, ensuring a comprehensive foundation for our generation process.
Model Development
We fine-tuned the ProtT5 model using LoRA (Low-Rank Adaptation) Adapters to adapt it specifically for our task of generating peptide sequences. This approach allows the model to learn patterns from our curated dataset of peptides and their target serotypes.
The fine-tuning process enables the model to understand the relationship between peptide structures and their biological targets, creating a specialized tool for antiviral peptide generation.
Inhibition Prediction
To predict the effectiveness of generated peptides, we implemented machine learning models for regression analysis. Embeddings generated from the ProtT5 model's encoder serve as numerical input features for these predictive models.
The Random Forest regression model was selected to predict inhibition scores, where lower values (similar to IC50 measurements) indicate greater potency and effectiveness against target viruses.
Molecular Docking
This critical phase involved analyzing the generated peptides' interaction with the DENV E protein using AutoDock Vina. The process required obtaining 3D structures of both the receptor (E Protein) and ligands (Peptides).
We utilized tools like the PepFOLD server for structure prediction and the Obabel library for format conversion to PDBQT. AutoDock Vina simulates various binding positions and orientations, predicting the optimal binding pose and affinity.
ADMET Properties
To assess the generated peptides' effectiveness and safety profile, we predicted their ADMET properties (Absorption, Distribution, Metabolism, Excretion, and Toxicity) using the pkCSM server.
Peptide sequences were converted to SMILES format using NovoPro Labs Tools, enabling comprehensive pharmacokinetic and toxicological analysis to ensure the viability of our designed peptides as potential therapeutic agents.
My Scientific Journey
As an intern at Kinase AI, I have learned things beyond my college curriculum. My interest in scientific discovery led me to grab this opportunity to experience this field that I am curious about. A little FYI, before entering college as a STEM student, I loved unraveling chemical processes, how these tiny molecules interact to form cells, and how those cells build the complex systems that make up humans, plants, and animals.
In my time in this internship, I have explored the structure of a virus, the proteins, how antibodies work, and aside from peptide that usually being used on skincare, I discovered that it is not limited to being an ingredient to my daily routine. Kidding aside, the dengue virus is a huge threat worldwide, and being able to contribute even a small piece toward understanding or combating it was both humbling and exciting.